For many new Ansible-based projects, I build my tests in Molecule, so I can easily run them locally or in CI. I also started using GitHub Actions for many of my new Ansible projects, just because it's so easy to get started and integrate with GitHub repositories.
I'm actually going to talk about this strategy in my next Ansible 101 live stream, covering Testing Ansible playbooks with Molecule and GitHub Actions CI, but I also wanted to highlight one thing that helps me when reviewing or observing playbook and molecule output, and that's color.
By default, in an interactive terminal session, Ansible colorizes its output so failures get 'red' color, good things / ok gets 'green', and changes get 'yellow-ish'. Also, warnings get a magenta color, which flags them well so you can go and fix them as soon as possible (that's one core principle I advocate to make your playbooks maintainable and scalable).
But by default, in a CI environment like GitHub Actions, there is no interactive TTY, so Ansible doesn't output color codes. Molecule doesn't either, since it's following standard convention, and as a result, when you review the logs for your workflow run, you see output like this:
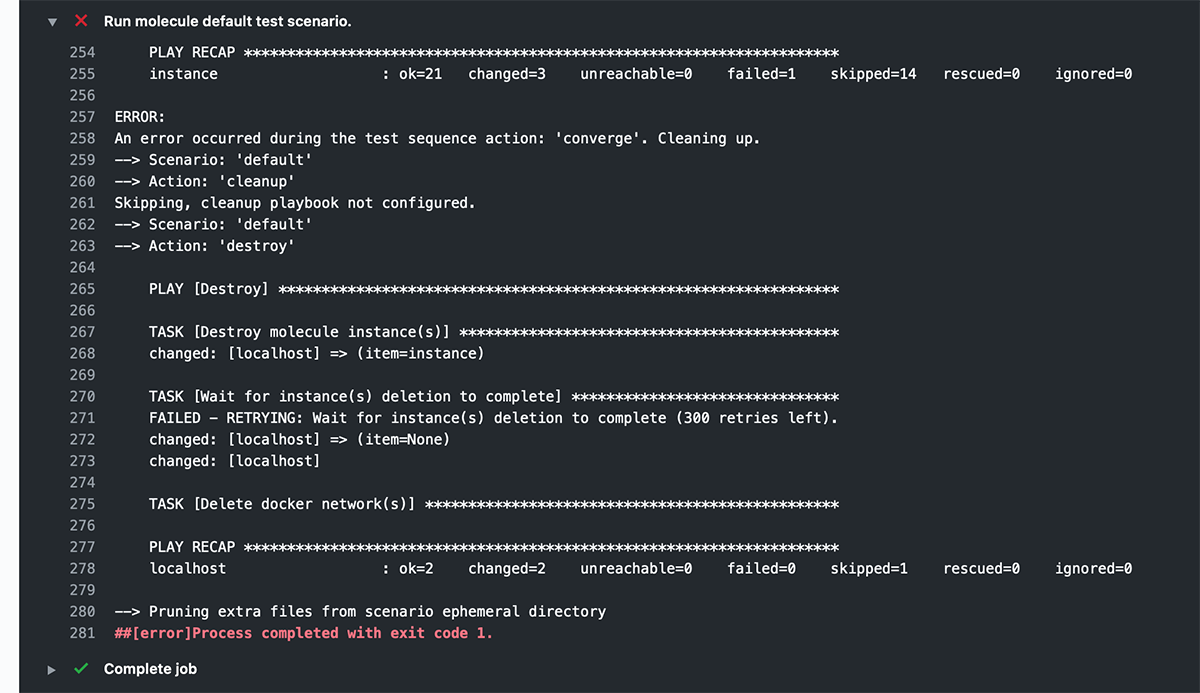
This is from the following build step in a GitHub Actions job:
- name: Run Molecule tests.
run: molecule testThere are two different environment variables you need to pass—one for Molecule, and one for Ansible—to make the output pretty and colorful, so it's easier to glance through a test run's output:
PY_COLORS: This is a somewhat standard way to tell Python-based tools to force color output, even without TTY. This forces Molecule to generate colorized output, but not Ansible.ANSIBLE_FORCE_COLOR: This forces Ansible to generate colorized output.
So, add these two variables to the GitHub Actions step in the env:
- name: Run Molecule tests.
run: molecule test
env:
PY_COLORS: '1'
ANSIBLE_FORCE_COLOR: '1'And now, after a workflow run, you get pretty colors!
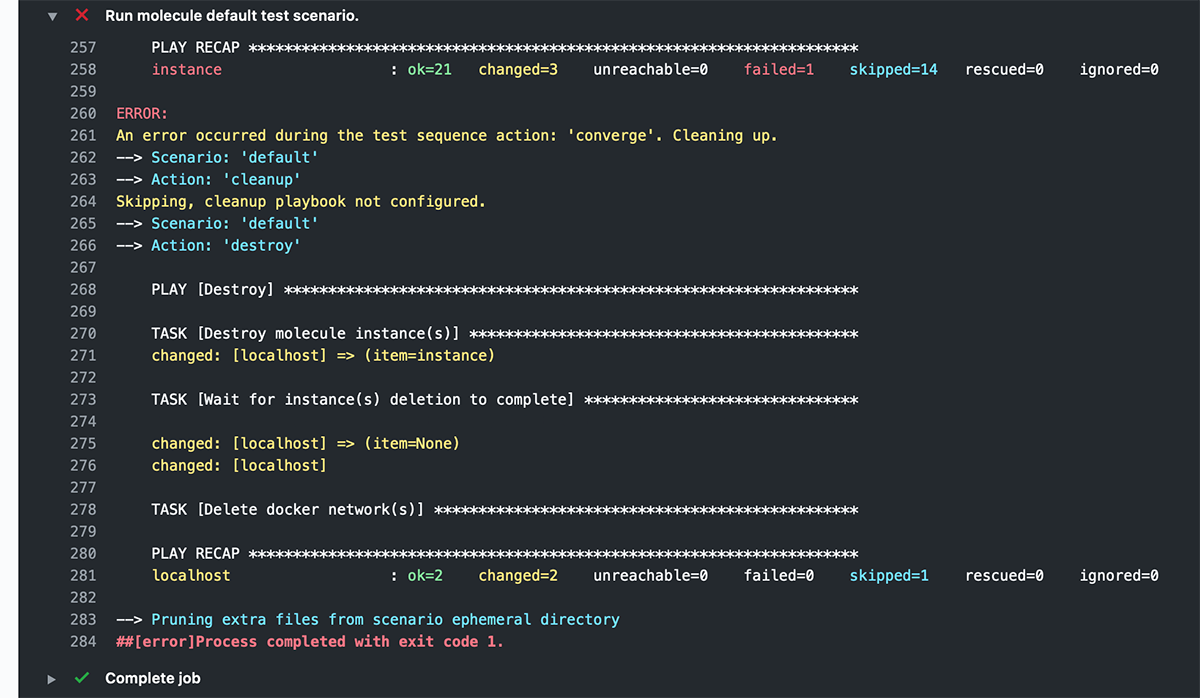
Learn more about running GitHub Actions workflows to test your Ansible content with Molecule in chapter 12 of Ansible for DevOps.
Comments
You can also run it from the jobs environment.
- { "distro":"centos-7", "command":"/usr/sbin/init" }- { "distro":"centos-8", "command":"/usr/sbin/init" }
- { "distro":"fedora-32", "command":"/usr/sbin/init" }
- { "distro":"fedora-31", "command":"/usr/sbin/init" }
- { "distro":"fedora-30", "command":"/usr/lib/systemd/systemd" }
- { "distro":"ubuntu-16.04", "command":"/sbin/init" }
- { "distro":"ubuntu-18.04", "command":"/lib/systemd/systemd" }
- { "distro":"ubuntu-20.04", "command":"/lib/systemd/systemd" }
- { "distro":"debian-9", "command":"/lib/systemd/systemd" }
- { "distro":"debian-10", "command":"/lib/systemd/systemd" }
If someone is searching how to do this with Gitlab, just add following to gitlab-ci.yml
variables:ANSIBLE_FORCE_COLOR: 'true'
PY_COLORS: '1'
For ansible-test, the ANSIBLE_FORCE_COLOR: '1' env var is not passed and we must add the "--color" parameter as answered to your own bug. It seemed to me it has its place here :)
https://github.com/ansible/ansible/issues/71885